

Echinoderm model systems, homology, and phylogenetic inference: Comment and reply to Paul (2021)
JENNIFER E. BAUER, SARAH L. SHEFFIELD, JOHNNY A. WATERS, and COLIN D. SUMRALL
Understanding the phylogenetic relationship among derived blastozoans has been a goal of researchers since phylogenetic methodologies were first applied to Paleozoic echinoderms. Paul (2021) proposed a new “pan-dichoporites” group to circumscribe early Paleozoic blastozoans. Unfortunately, this work includes many inaccuracies, non-reproducible analyses, and nonstandard method choices that confuse rather than advance the understanding of echinoderm paleobiology. Herein, we focus on key aspects of philosophy, methodology, and data reproducibility the publication of Paul (2021) raises that need to be addressed and considered by echinoderm researchers as they assess the concept of pan-dichoporite echinoderms.
The phylogenetic concept of Echinodermata was defined (Sumrall 2020a) as the crown clade originating with the most recent common ancestor of Asterias rubens Linnaeus, 1758 (Asteroidea); Ophiura ophiura Linnaeus, 1758 (Ophiuroidea); Strongylocentrotus purpuratus (Stimpson, 1857) (Echinoidea); Cucumaria frondosa (Gunnerus, 1767) (Holothuroidea), Metacrinus rotundus Carpenter, 1885 (Crinoidea); and Xyloplax medusiformis Baker, Rowe, and Clark 1986 (Concentricycloidea). As defined, this clade circumscribes five extant groups nested in two clades. Eleutherozoa includes Asteroidea (including Xyloplax see Janies et al. 2011), Ophiuroidea, Holothuroidea, and Echinoidea. Crinoidea forms a separate clade. In contrast, there are more than 20 distinct echinoderm clades that are extinct. Among extant groups, Echinoidea is often used as a model system to describe all Echinodermata despite having extremely high rates of missing data because of its highly apomorphic morphologies in echinoderm-wide morphological datasets (see Deline 2021). Echinoids diverged from their closest relatives (Holothuroidea) more than 450 million years ago with crown echinoids originating in the Permian (Thompson et al. 2017).
To better understand body-wall homologies, common ontogenetic patterns, major events in body plan evolution, and the identification of synapomorphies among morphologically disparate echinoderm clades, Mooi et al. (1994) proposed the Extraxial-Axial Theory (EAT) largely based on a novel understanding of larval development in extant echinoids. A discussion of the validity of EAT is outside the scope of this commentary, but its use by Paul (2021) does raise important philosophical questions. Are eleutherozoans, including echinoids, an appropriate model to understand morphologically diverse extinct echinoderm clades such as the blastozoans and other pelmatozoans?
Assumptions that Paleozoic echinoderms would have had similar developmental pathways to extant echinoids are problematic. At present, very little information exists on the larval histories of Paleozoic pelmatozoans (Sumrall and Sprinkle 1998; Sevastopulo 2005). This lack of developmental data should be mentioned as a caveat in all studies invoking EAT as a model for homology among non-eleutherozoan taxa. For example, ocular plate rule (OPR) and radial water vessels (RWV) are concepts based on extant eleutherozoans, but ocular plates are documented only in echinoids and asteroids and do not occur in other groups.
Universal Elemental Homology (UEH) is a homology hypothesis that takes into account comparative anatomy, ontogeny, function, and position to identify homologous plates across taxa (Sumrall 2010; Sumrall and Waters 2012). EAT and UEH are two different schemes for understanding homology that should not be considered mutually exclusive. Sumrall and Waters (2012: 956) attempted to underscore this point with the following: “The EAT theory has been useful for understanding homology at the highest taxonomic levels where deep structure is illuminated by these regional homologies. Universal elemental homology (described here for stemmed echinoderms) takes the understanding of homology to the next level by allowing the identification, in many cases, of individual plates across clades. Thus, evolutionary changes in shape or plate contact relationships can be used to generate characters that are useful for reconstructing phylogeny at the lowest taxonomic levels.”
The plate nomenclature update from Sumrall and Waters (2012) was useful in showcasing the critical issue with plate naming systems in echinoderm paleobiology. The work by Paul (2021) seemingly reproduces a version of this update but following the EAT schema. Paul (2021) largely ignores the efforts to understand the homologous oral plates in blastozoans in the UEH schema outside of stating they are incongruent with EAT (Sumrall and Waters 2012). We completely understand that authors may not fully agree with UEH; however, when disagreement arises between homology schemes a case should be presented to showcase the efficacy of one approach as better than the other rather than to completely disregard the body of published work.
Similar to the issues that arise with applying EAT to blastozoans, UEH does not work when applied to echinoids. The homologous skeletal elements outlined in UEH do not exist in echinoids, which would all be rendered as non-applicable data in morphological and phylogenetic downstream analyses. Therefore, the homology scheme employed to assess and generate character state data for the echinoderm group is important in understanding small (e.g., Blastoidea) and large group (e.g., Blastozoa) evolutionary patterns. Our hypotheses of homology must be critically evaluated, as phylogenetic inferences are sensitive to these data and can lead us to erroneous understandings of evolutionary relationships.
Abbreviations.—EAT, Extraxial-Axial Theory; UEH, Universal Elemental Homology.
Data inputs and phylogenetic inference
Phylogenetic tools are powerful, but the output is highly dependent on the data input. For reproducibility, all published studies should include supplemental files of all files used to perform the analysis. Paul (2021) stated that he only used early Paleozoic blastozoans, though the eublastoid used in this analysis is a Carboniferous genus when there are Silurian taxa that would have been better suited (e.g., Polydeltoideus or Troosticrinus). There is also an inaccuracy listed in Table 1 with the placement of Macurdablastus in Stephanocrinidae. This taxonomic placement is not supported by previous work (e.g., Broadhead 1984; Bodenbender and Fisher 2001; Bauer et al. 2019).
Phylogenetic characters are assumed to be both heritable and independent of one another. These assumptions require the removal of character sets that break these assumptions such as ecology and stratigraphy (Sumrall 1997). Swofford and Olsen (1990) present the challenges that would arise in computing phylogenetic trees if dependent characters were used. In Paul (2021), no character explanations were provided to clarify the thought behind character constructions, making it difficult to ascertain the morphologies encompassed by each character state. However, many characters used in the analysis appear to be constructed outside of these assumptions or use arbitrary ranges that do not describe alternate expressions of homology (Weins 2001; Wiley and Lieberman 2011). Many characters are also coded incorrectly. For example, character 11 is “radials being present or absent”. Those with missing radials (e.g., Thomatocystis) should then be coded as non-applicable in other characters involving radial plates, though they are in Paul (2021) as a “?”. Three of the fourteen taxa are missing more than 10 of the 24 characters (42%). Missing data has a large effect on tree topology, especially in such a small dataset. This is critical because character selection (and taxon selection) can have large influences on tree outputs (Wiley and Lieberman 2011).
For any phylogenetic analysis, it is commonplace for character descriptions and codings to be publicly available, allowing readers to follow and understand the author’s justifications for character description and character state transformation selections. Missing character data can artificially alter tree topologies as it relates to accuracy and support (Scotland et al. 2003). Two taxa in the work of Paul (2021) are missing 11 and 12 characters. This, in concert with the three parsimony-uninformative characters, problematically leaves fewer than 50% of the characters scored for these taxa, possibly resulting in a decrease in accuracy of reconstructed trees (Huelsenbeck 1991; Hartmann and Vision 2008). With too much information missing, maximum parsimony will recover many alternate topologies and the consensus summary trees may be misleading (Wilkinson 1995). Best practices suggest that results are more resolved when there are at least two to three times the characters than there are taxa (Scotland et al. 2003).
Phylogeny.—General methodological and interpretive errors exist in the recent Paul (2021) work. For example, there is a rather lengthy discussion regarding the placement of the unusual glyptocystitoid Rhombifera in the blastozoan evolutionary history. The author suggests that inclusive blastoids (i.e., eublastoids, coronoids, Lysocystites, Macurdablastus) are most closely related to Rhombifera (Paul 2021: 48). However, the results of the phylogenetic analysis (Paul 2021: 59) do not align with this assessment as Rhombifera is not recovered as the sister group to the inclusive Blastoidea. There is no discussion on why Rhombifera is not sister taxa to the inclusive Blastoidea as one would expect given the earlier sections. The author concludes that the ambulacrals of Lysocystites are Rhombifera radials and eublastoid lancets. Paul (2021) does not discuss that if this is the case, we would expect to see ambulacrals on Thomacystis and Caryocrinites as well, given the results of his phylogeny. The phylogenetic analysis performed here used a heuristic search method for 14 taxa. However, best practices in phylogenetics suggest that a heuristic search for this particular analysis is inappropriate, as the size of the dataset allows for the use of exact methods (i.e., exhaustive searches or branch and bound searches; Swofford and Olsen 1990).
Support indices are useful measures that allow researchers to gain information on the dataset and resulting tree topology, but they provide little information about support for monophyletic groups within the tree (Wiley and Lieberman 2011). Other measures of nodal support (i.e., bootstrap) that resample the character matrix would be more valuable here. The author describes 100% support in reference to the 50% majority rule consensus tree but this information indicates that of the six resulting trees the relationships were recovered 100% of the time. It is not indicating that the nodes are 100% supported. Upon request, the PAUP nexus and log files were sent for re-analysis. Our intention was not to correct the character coding dataset, rather to illustrate pathways for improvement. We were able to replicate the exact results from PAUP for the parsimony analysis. With this tree topology, we ran a bootstrap analysis, resampling all 24 characters for 100 replicates to determine nodal support. Only six of the 11 nodes had ≥ 50% support. Unsurprisingly, inclusive Blastoidea is found at 79%; Caryocrinites + Thomacystis found at 76%; the large grouping of all taxa except Akadocrinus and Cambrocrinus is at 72%. Support for Macurdablastus and Codaster is at 68%. Lysocystites and Stephanocrinus is at 65%. Finally, the large grouping of Macrocystella upward is supported at 50% (Fig. 1A). Since parsimony analysis excludes any characters that are considered “parsimony uninformative”, this includes three characters of the 24, we also re-analyzed the dataset with maximum likelihood to see if utilizing all 24 characters with the mKv model (Lewis 2001) provided differing results. Three trees were retained with a LnL score of 150.4167 with six nodes which had ≥ 50% support (Fig. 1B). The major difference between the recovered trees is the placement of Ridersia, which is not surprising given the number of missing characters (8/24) for that taxon (Fig. 1). In this tree the support for Ridersia upward is much higher at 91% than in the parsimony results. The relationships between the inclusive Blastoidea match those proposed by Bauer et al. (2019) and the support is slightly higher than that of the parsimony tree at 81%. In this analysis, Rhombifera is always sister taxon to Lepadocystis, which was an unresolved grouping in Paul’s (2021) parsimony tree. Most notably, the taxon that is unstable across the three most likely trees (Ridersia) is not the taxon (Lepadocystis) that was unstable in the parsimony analysis (Fig. 1).
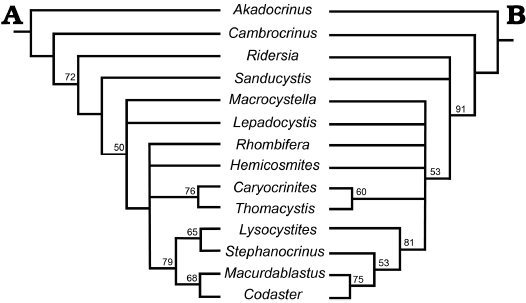
Fig. 1. Tree comparison between two phylogenetic inference methods with bootstrap support at the nodes. A. Phylogenetic hypothesis from Paul (2021) inferred via maximum parsimony. B. Phylogenetic hypothesis inferred via maximum likelihood.
The pan-naming convention in Paul (2021) is used incorrectly for the proposed pan-dichoporites. As laid out in the PhyloCode (Cantino and De Queiroz 2020), pan groups are designated as total groups that include the related name-bearing crown groups. For example, Echinodermata is a crown clade including all descendants of the last common ancestor of Echinoidea, Asteroidea, Ophiuroidea, Holothuroidea and Crinoidea (Sumrall 2020a). This circumscribes a clade that includes all modern taxa and such fossil taxa that are descended from the most inclusive node. Sumrall (2020b) defined Pan-Echinodermata as the total group echinodermata; that is, the stem lineage that includes all taxa closer to Echinodermata than to any other crown. This effectively aligns with the traditional non-phylogenetic diagnosed Echinodermata including stylophorans and other basal taxa. The pan-naming convention simply cannot be used for extinct lineages. There is no crown group within blastozoans as the last known members were extinct by the end of the Permian. Likewise, there is no total group in the blastozoans because there is no set of taxa closer to an non-extant crown lineage than another crown lineage. Pan-dichoporites is effectively a synonym of Blastozoa and while they were not formally defined either by PhyloCode rules or by ZooBank, names for several derived echinoderm clade names were previously suggested by Sumrall (1997).
Stratigraphy.—Stratigraphy has played an interesting role in understanding echinoderm evolutionary relationships. The concept of stratocladistics (Bodenbender and Fisher 2001) was established using blastoids. However, stratigraphic information should not be incorporated into a phylogenetic character matrix (Sumrall 1997); it is information that is unrelated to the heritable traits of the animals. However, it can be incorporated into other aspects of phylogenetic analysis as a parameter to better understand and analyze clades of interest. On several occasions the author makes targeted remarks regarding the stratigraphic occurrences of fossils and the evolution of these groups. These statements are problematic in different ways. The first reads, “Both the suggestions that Hemicosmites preceded the glyptocystitoids and that Lysocystites preceded the remaining blastoids sensu lato are counter to known stratigraphy of occurrences” (p. 59). This frame of thinking excludes considerations of evolutionary processes and is not in terms of the most recent common ancestor but instead only of the terminal taxa. These thoughts are exclusive and can be confusing to readers. In another section the author writes, “It has become fashionable to ignore stratigraphy and the grounds of the incompleteness of the fossil record” (p. 59). This is ignoring the recent and large body of work utilizing stratigraphy in concert with phylogeny in fossil invertebrates (e.g., Congreve et al. 2019; Lam et al. 2018, 2021; Bauer 2021). Stratigraphy is not being ignored, rather it is more fully and appropriately being utilized. Just because a taxon is stratigraphically older does not mean it is ancestral.
Acknowledgements.—We thank Maggie Limbeck (University of Tennessee Knoxville), Whitney Lapic and Stephen Hill (both University of South Florida) for comments and discussions on earlier versions of this commentary.
References
Baker, A.N., Rowe, F.W.E., and Clark, H.E.S. 1986.) A new class of echinodermata from New Zealand. Nature 321: 862–864. Crossref
Bauer, J.E. 2021. Paleobiogeography, paleoecology, diversity, and speciation patterns in the Eublastoidea (Blastozoa: Echinodermata). Paleobiology 47: 221–235. Crossref
Bauer, J.E., Waters, J.A., and Sumrall, C.D. 2019. Redescription of Macurdablastus and redefinition of Eublastoidea as a clade of Blastoidea (Echinodermata). Palaeontology 62: 1003–1013. Crossref
Bodenbender, B.E. and Fisher, D.C. 2001. Stratocladistic analysis of blastoid phylogeny. Journal of Paleontology 75: 351–369. Crossref
Broadhead, T.W. 1984. Macurdablastus, a Middle Ordovician blastoid from the southern Appalachians. The University of Kansas Paleontological Contributions 110: 1–9.
Cantino, P.D. and de Queiroz, K. 2020. PhyloCode: A Phylogenetic Code of Biological Nomenclature. Available from https://www.ohio.edu/PhyloCode/PhyloCode.pdf [accessed 1 November 2021].
Congreve, C.R., Krug, A.Z., and Patzkowsky, M.E. 2019. Evolutionary and biogeographical shifts in response to the Late Ordovician mass extinction. Palaeontology 62: 267–285. Crossref
Deline, B. 2021. Echinoderm Morphological Disparity: Methods, Patterns, and Possibilities. Elements of Paleontology. 40 pp. Cambridge University Press, Cambridge. Crossref
Hartmann, S. and Vision, T.J. 2008. Using ESTs for phylogenomics: can one accurately infer a phylogenetic tree from a gappy alignment? BMC Evolutionary Biology 8: 1–13. Crossref
Huelsenbeck, J.P. 1991. When are fossils better than extant taxa in phylogenetic analysis? Systematic Biology 40: 458–469. Crossref
Janies, D.A., Voight, J.R., and Daly, M. 2011. Echinoderm phylogeny including Xyloplax, progenetic asteroid. Systematic Biology 60: 420–438. Crossref
Lam, A.R., Sheffield, S.L., and Matzke, N.J. 2021. Estimating dispersal and evolutionary dynamics in diploporan blastozoans (Echinodermata) across the great Ordovician biodiversification event. Paleobiology 47: 198–220. Crossref
Lam, A.R., Stigall, A.L., and Matzke, N.J. 2018. Dispersal in the Ordovician: speciation patterns and paleobiogeographic analyses of brachiopods and trilobites. Palaeogeography, Palaeoclimatology, Palaeoecology 489: 147–165. Crossref
Lewis, P.O. 2001. A likelihood approach to estimating phylogeny from discrete morphological character data. Systematic Biology 50: 913–925. Crossref
Mongiardino Koch, N. and Thompson, J.R. 2021. A total-evidence dated phylogeny of Echinoidea combining phylogenomic and paleontological data. Systematic Biology 70: 421–439. Crossref
Mooi, R., David, B., and Marchand, D. 1994. Echinoderm skeletal homologies: classical morphology meets modern phylogenetics. In: B. David, A. Guille, J.P. Féral, and M. Roux (eds.), Echinoderms Through Time (Echinoderms Dijon), 87–95. Balkema, Rotterdam. Crossref
Paul, C.R.C 2021. New insights into the origin and relationships of blastoid echinoderms. Acta Palaeontologica Polonica 66: 41–62. Crossref
Paul, C.R.C. and Smith, A.B. 1984. The early radiation and phylogeny of echinoderms. Biological Reviews 59: 443–481. Crossref
Scotland, R.W., Olmstead, R.G., and Bennett, J.R. 2003. Phylogeny reconstruction: the role of morphology. Systematic Biology 52: 539–548. Crossref
Sevastopulo, G.D. 2005. The early ontogeny of blastoids. Geological Journal 40: 351–362. Crossref
Sprinkle, J. 1973. Morphology and Evolution of Blastozoan Echinoderms. 284 pp. Museum of Comparative Zoology, Harvard University, Cambridge. Crossref
Sumrall, C.D. 1997. The role of fossils in the phylogenetic reconstruction of Echinodermata. The Paleontological Society Papers 3: 267–288. Crossref
Sumrall, C.D. 2010. A model for elemental homology for the peristome and ambulacra in blastozoan echinoderms. In: L.G. Harris, S.A. Böttger, C.W. Walker, and M.P. Lesser (eds.), Echinoderms: Durham, 269–276. Taylor & Francis, London.
Sumrall, C.D. and Sprinkle, J. 1998. Early ontogeny of the glyptocystitid rhombiferan Lepadocystis moorei. In: M.D.C. Carnevalli and F. Bonasoro (eds.), Echinoderm Research 1998. Proceedings of the Fifth European Conference on Echinoderms, 409–414. A.A. Balkema, Rotterdam.
Sumrall, C.D. and Waters, J.A. 2012. Universal elemental homology in glyptocystitoids, hemicosmitoids, coronoids and blastoids: steps to- wards echinoderm phylogenetic reconstruction in derived Blastozoa. Journal of Paleontology 86: 956–972. Crossref
Sumrall, C.D. 2020a. Echinodermata. In: K. de Queiroz, J. Gauthier, and P. Cantino (eds.), Phylonyms: A Companion Volume to the PhyloCode, 645–648. Taylor & Francis Group, London.
Sumrall, C.D. 2020b. Pan-Echinodermata. In: K. de Queiroz, J. Gauthier, and P. Cantino (eds.), Phylonyms: A Companion Volume to the PhyloCode, 641–644. Taylor & Francis Group, London.
Swofford, D.L. and Olsen, G.J. 1990. Phylogeny reconstruction. In: D.M. Hillis and C. Moritz (eds.), Molecular Systematics, 411–501. Sinauer Associates, Sunderland.
Thompson, J.R., Petsios, E., and Bottjer, D.J. 2017. A diverse assemblage of Permian echinoids (Echinodermata, Echinoidea) and implications for character evolution in early crown group echinoids. Journal of Paleontology 91: 767–780. Crossref
Wiens, J.J. 2001. Character analysis in morphological phylogenetics: problems and solutions. Systematic Biology 50: 689–699. Crossref
Wiley, E.O., and Lieberman, B.S. 2011. Phylogenetics: Theory and Practice of Phylogenetic Systematics. 432 pp. John Wiley & Sons, New Jersey. Crossref
Wilkinson, M. 1995. Coping with abundant missing entries in phylogenetic inference using parsimony. Systematic Biology 44: 501–514. Crossref
Jennifer E. Bauer [bauerjen@umich.edu], University of Michigan Museum of Paleontology, 1105 North University Ave., Ann Arbor, MI 48109 USA.
Sarah L. Sheffield [ssheffield2@usf.edu], 4202 E. Fowler Ave, NES 102, Tampa FL, 33620, USA.
Johnny A. Waters [watersja@appstate.edu], Department of Geological and Environmental Sciences, Appalachian State University, 033 Rankin Science West, ASU Box 32067 Boone, NC 28608-2067, USA.
Colin D. Sumrall [csumrall@utk.edu], Department of Earth and Planetary Sciences, The University of Tennessee, 1621 Cumberland Avenue, 602 Strong Hall, Knoxville, TN 37996-1526, USA.
Received 2 November 2021, accepted 19 November 2021, available online 15 February 2022.
Copyright © 2022 J.E. Bauer et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License (for details please see http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Acta Palaeontol. Pol. 67 (2): 465–468, 2022
https://doi.org/10.4202/app.00956.2021